Windowed reading and writing
Beginning in rasterio 0.3, you can read and write “windows” of raster files. This feature allows you to work on rasters that are larger than your computers RAM or process chunks of large rasters in parallel.
Windows
A Window is a view onto a rectangular subset of a raster dataset and is
described in rasterio by column and row offsets and width and height
in pixels. These may be ints or floats.
from rasterio.windows import Window
Window(col_off, row_off, width, height)
Windows may also be constructed from numpy array index tuples or slice objects. Only int values are permitted in these cases.
Window.from_slices((row_start, row_stop), (col_start, col_stop))
Window.from_slices(slice(row_start, row_stop), slice(col_start, col_stop))
If height and width keyword arguments are passed to from_slices(), relative
and open-ended slices may be used.
Window.from_slices(slice(None), slice(None), height=100, width=100)
# Window(col_off=0.0, row_off=0.0, width=100.0, height=100.0)
Window.from_slices(slice(10, -10), slice(10, -10), height=100, width=100)
# Window(col_off=10, row_off=10, width=80, height=80)
Reading
Here is an example of reading a 256 row x 512 column subset of the rasterio test file.
>>> import rasterio
>>> with rasterio.open('tests/data/RGB.byte.tif') as src:
... w = src.read(1, window=Window(0, 0, 512, 256))
...
>>> print(w.shape)
(256, 512)
Attention
In getting data to fill a window Rasterio will read the entirety of one or more chunks of data from the dataset. If you’re reading from a GeoTIFF with 512 x 512 pixel chunks (blocks), that determines the minimum number of bytes that will be read from disk or copied over your network, even if your read window is only 1 x 1 pixels. In the case that your source dataset does not use chunks (rare, but possible) Rasterio will read the entire dataset in order to fill even a 1 x 1 pixel window. In practice, it’s important to chunk the data you create and store for your applications.
Writing
Writing works similarly. The following creates a blank 500 column x 300 row GeoTIFF and plops 37,500 pixels with value 127 into a window 30 pixels down from and 50 pixels to the right of the upper left corner of the GeoTIFF.
image = numpy.ones((150, 250), dtype=rasterio.ubyte) * 127
with rasterio.open(
'/tmp/example.tif', 'w',
driver='GTiff', width=500, height=300, count=1,
dtype=image.dtype) as dst:
dst.write(image, window=Window(50, 30, 250, 150), indexes=1)
The result:
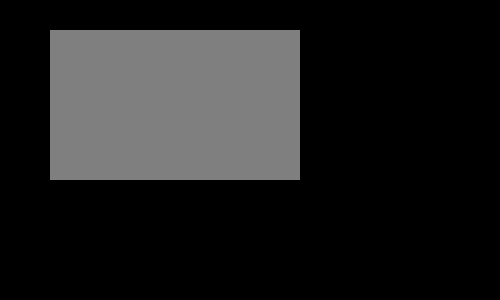
Decimation
If the write window is smaller than the data, the data will be decimated. Below, the window is scaled to one third of the source image.
with rasterio.open('tests/data/RGB.byte.tif') as src:
b, g, r = (src.read(k) for k in (1, 2, 3))
# src.height = 718, src.width = 791
write_window = Window.from_slices((30, 269), (50, 313))
# write_window.height = 239, write_window.width = 263
with rasterio.open(
'/tmp/example.tif', 'w',
driver='GTiff', width=500, height=300, count=3,
dtype=r.dtype) as dst:
for k, arr in [(1, b), (2, g), (3, r)]:
dst.write(arr, indexes=k, window=write_window)
And the result:

Data windows
Sometimes it is desirable to crop off an outer boundary of NODATA values around
a dataset. You can do this with get_data_window():
from rasterio.windows import get_data_window
with rasterio.open('tests/data/RGB.byte.tif') as src:
window = get_data_window(src.read(1, masked=True))
# window = Window(col_off=13, row_off=3, width=757, height=711)
kwargs = src.meta.copy()
kwargs.update({
'height': window.height,
'width': window.width,
'transform': rasterio.windows.transform(window, src.transform)})
with rasterio.open('/tmp/cropped.tif', 'w', **kwargs) as dst:
dst.write(src.read(window=window))
Window transforms
The affine transform of a window can be accessed using a dataset’s
window_transform() method:
>>> import rasterio
>>> from rasterio.windows import Window
>>> win = Window(256, 256, 128, 128)
>>> with rasterio.open('tests/data/RGB.byte.tif') as src:
... src_transform = src.transform
... win_transform = src.window_transform(win)
...
>>> print(src_transform)
| 300.04, 0.00, 101985.00|
| 0.00,-300.04, 2826915.00|
| 0.00, 0.00, 1.00|
>>> print(win_transform)
| 300.04, 0.00, 178794.71|
| 0.00,-300.04, 2750104.30|
| 0.00, 0.00, 1.00|
Window utilities
Basic union and intersection operations are available for windows, to streamline operations across dynamically created windows for a series of bands or datasets with the same full extent.
>>> from rasterio import windows
>>> # Full window is ((0, 1000), (0, 500))
>>> window1 = Window(10, 100, 490, 400)
>>> window2 = Window(50, 10, 200, 140)
>>> windows.union(window1, window2)
Window(col_off=10, row_off=10, width=490, height=490)
>>> windows.intersection(window1, window2)
Window(col_off=50, row_off=100, width=200, height=50)
Blocks
Raster datasets are generally composed of multiple blocks of data and windowed reads and writes are most efficient when the windows match the dataset’s own block structure. When a file is opened to read, the shape of blocks for any band can be had from the block_shapes property.
>>> with rasterio.open('tests/data/RGB.byte.tif') as src:
... for i, shape in enumerate(src.block_shapes, 1):
... print((i, shape))
...
(1, (3, 791))
(2, (3, 791))
(3, (3, 791))
The block windows themselves can be had from the block_windows function.
>>> with rasterio.open('tests/data/RGB.byte.tif') as src:
... for ji, window in src.block_windows(1):
... print((ji, window))
...
((0, 0), ((0, 3), (0, 791)))
((1, 0), ((3, 6), (0, 791)))
...
This function returns an iterator that yields a pair of values. The second is
a window tuple that can be used in calls to read()
or write(). The first is the pair of row and column
indexes of this block within all blocks of the dataset.
You may read windows of data from a file block-by-block like this.
>>> with rasterio.open('tests/data/RGB.byte.tif') as src:
... for ji, window in src.block_windows(1):
... r = src.read(1, window=window)
... print(r.shape)
... break
...
(3, 791)
Well-bred files have identically blocked bands, but GDAL allows otherwise and it’s a good idea to test this assumption in your code.
>>> with rasterio.open('tests/data/RGB.byte.tif') as src:
... assert len(set(src.block_shapes)) == 1
... for ji, window in src.block_windows(1):
... b, g, r = (src.read(k, window=window) for k in (1, 2, 3))
... print((ji, r.shape, g.shape, b.shape))
... break
...
((0, 0), (3, 791), (3, 791), (3, 791))
The block_shapes property is a band-ordered list of block shapes and set(src.block_shapes) gives you the set of unique shapes. Asserting that there is only one item in the set is effectively the same as asserting that all bands have the same block structure. If they do, you can use the same windows for each.