Reading Datasets
Todo
- Discuss and/or link to topics
supported formats, drivers
vsi
tags
profile
crs
transforms
dtypes
Dataset objects provide read, read-write, and write access to raster data files
and are obtained by calling rasterio.open(). That function mimics Python’s
built-in open() and the dataset objects it returns mimic Python file
objects.
>>> import rasterio
>>> src = rasterio.open('tests/data/RGB.byte.tif')
>>> src
<open DatasetReader name='tests/data/RGB.byte.tif' mode='r'>
>>> src.name
'tests/data/RGB.byte.tif'
>>> src.mode
'r'
>>> src.closed
False
If you try to access a nonexistent path, rasterio.open() does the same
thing as open(), raising an exception immediately.
>>> open('/lol/wut.tif')
Traceback (most recent call last):
...
FileNotFoundError: [Errno 2] No such file or directory: '/lol/wut.tif'
>>> rasterio.open('/lol/wut.tif')
Traceback (most recent call last):
...
rasterio.errors.RasterioIOError: No such file or directory
Datasets generally have one or more bands (or layers). Following the GDAL convention, these are indexed starting with the number 1. The first band of a file can be read like this:
>>> array = src.read(1)
>>> array.shape
(718, 791)
The returned object is a 2-dimensional numpy.ndarray. The representation of
that array at the Python prompt is a summary; the GeoTIFF file that
Rasterio uses for testing has 0 values in the corners, but has nonzero values
elsewhere.
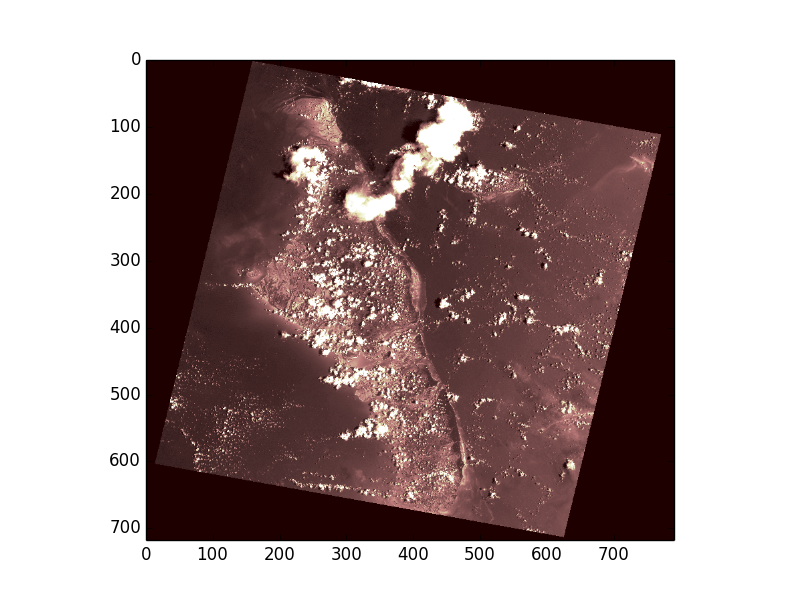
>>> from matplotlib import pyplot
>>> pyplot.imshow(array, cmap='pink')
<matplotlib.image.AxesImage object at 0x...>
>>> pyplot.show()
Instead of reading single bands, all bands of the input dataset can be read into
a 3-dimensonal ndarray. Note that the interpretation of the 3 axes is
(bands, rows, columns). See
Image processing software for more details on how to convert to the ordering expected by
some software.
>>> array = src.read()
>>> array.shape
(3, 718, 791)
In order to read smaller chunks of the dataset, refer to Windowed reading and writing.
The indexes, Numpy data types, and nodata values of all a dataset’s bands can
be had from its indexes, dtypes,
and nodatavals attributes.
>>> for i, dtype, nodataval in zip(src.indexes, src.dtypes, src.nodatavals):
... print(i, dtype, nodataval)
...
1 uint8 0.0
2 uint8 0.0
3 uint8 0.0
To close a dataset, call its close() method.
>>> src.close()
>>> src
<closed DatasetReader name='tests/data/RGB.byte.tif' mode='r'>
After it’s closed, data can no longer be read.
>>> src.read(1)
Traceback (most recent call last):
...
ValueError: can't read closed raster file
This is the same behavior as Python’s file.
>>> f = open('README.rst')
>>> f.close()
>>> f.read()
Traceback (most recent call last):
...
ValueError: I/O operation on closed file.
As Python file objects can, Rasterio datasets can manage the entry into
and exit from runtime contexts created using a with statement. This
ensures that files are closed no matter what exceptions may be raised within
the the block.
>>> with rasterio.open('tests/data/RGB.byte.tif', 'r') as one:
... with rasterio.open('tests/data/RGB.byte.tif', 'r') as two:
... print(two)
... print(one)
... raise Exception("an error occurred")
...
<open DatasetReader name='tests/data/RGB.byte.tif' mode='r'>
<open DatasetReader name='tests/data/RGB.byte.tif' mode='r'>
Traceback (most recent call last):
File "<stdin>", line 5, in <module>
Exception: an error occurred
>>> print(two)
<closed DatasetReader name='tests/data/RGB.byte.tif' mode='r'>
>>> print(one)
<closed DatasetReader name='tests/data/RGB.byte.tif' mode='r'>
Format-specific dataset reading options may be passed as keyword arguments. For example, to turn off all types of GeoTIFF georeference except that within the TIFF file’s keys and tags, pass GEOREF_SOURCES=’INTERNAL’.
>>> with rasterio.open('tests/data/RGB.byte.tif', GEOREF_SOURCES='INTERNAL') as dataset:
... # .aux.xml, .tab, .tfw sidecar files will be ignored.